2025 Swiss TB Award
The winners are Dr Imre Gonda for "The mycobacterial ABC transporter IrtAB employs a membrane-facing crevice for siderophore-mediated iron uptake" and Dr Galo A. Goig for "Transmission as a Key Driver of Resistance to the New Tuberculosis Drugs".
Dr Imre Gonda et al.
The mycobacterial ABC transporter IrtAB employs a membrane-facing crevice for siderophore-mediated iron uptake
 Drug-resistant Mycobacterium tuberculosis is a major global health threat. New antibiotics are urgently needed—especially those that target the unique biology of this pathogen. At the Seeger Lab at the University of Zürich, we focus on mycobacterial membrane transporters, the gatekeepers of this deadly bacterium. In our recent study, published in Nature Communications (2025), we investigated the mycobactin-mediated iron uptake cycle, with a particular focus on how the inner membrane transporter IrtAB imports vital, mycobactin-bound ferric ions (Fe3+).
Drug-resistant Mycobacterium tuberculosis is a major global health threat. New antibiotics are urgently needed—especially those that target the unique biology of this pathogen. At the Seeger Lab at the University of Zürich, we focus on mycobacterial membrane transporters, the gatekeepers of this deadly bacterium. In our recent study, published in Nature Communications (2025), we investigated the mycobactin-mediated iron uptake cycle, with a particular focus on how the inner membrane transporter IrtAB imports vital, mycobactin-bound ferric ions (Fe3+).
In the human body, iron and other essential metal ions are tightly bound by host proteins to limit bacterial growth. To survive under these restrictive conditions, bacteria employ high-affinity metal-chelating molecules known as siderophores. In M. tuberculosis, these siderophores are either secreted (carboxymycobactin, cMBT) or membrane-bound (mycobactin, MBT). Mycobactins are synthesized in the cytoplasm and transported to the periplasm by MmpL4/5 and the corresponding MmpS4/5 proteins. However, the proteins responsible for the mycobactin passage across the outer membrane remain unidentified. Once iron-loaded, mycobactins are taken back up through the inner membrane by the ABC transporter IrtAB, which also releases usable iron into the cytoplasm by reducing ferric to ferrous iron (Fe3+ to Fe2+) with its siderophore interaction domain. Finally, the empty mycobactins are recirculated to scavenge more iron.
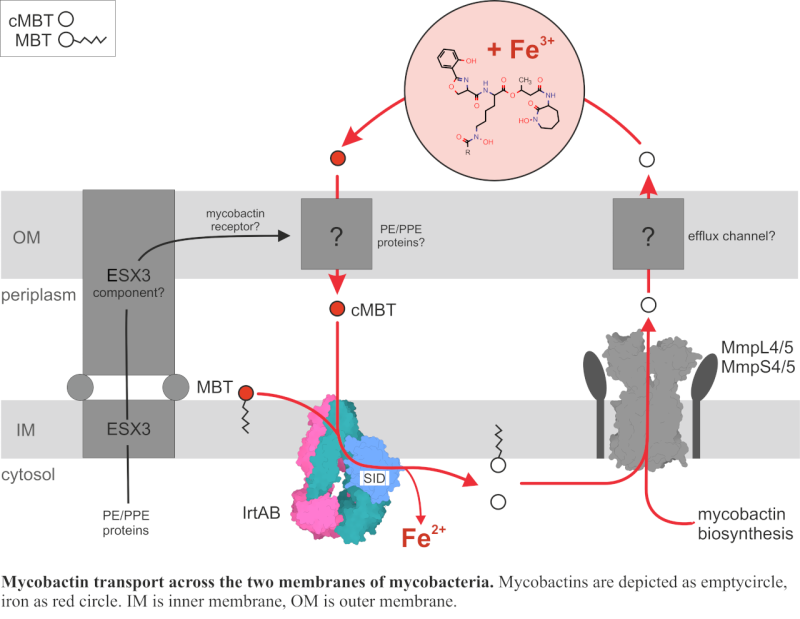
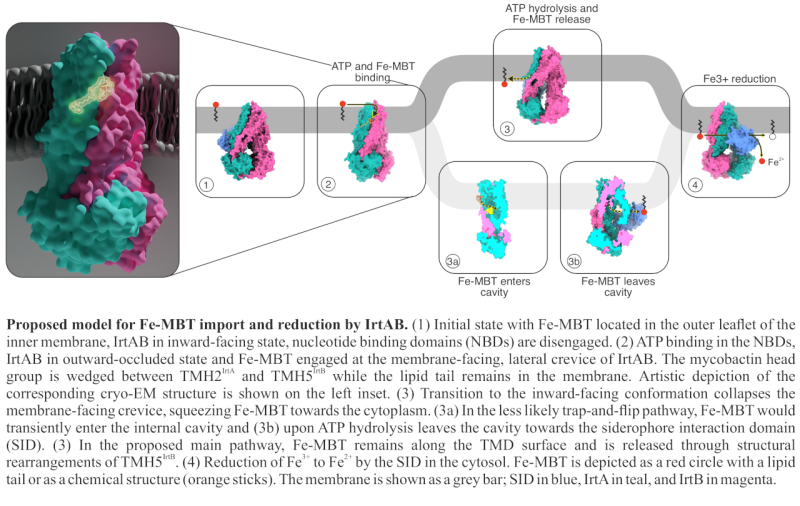
In our previous work, we provided functional and structural evidence, that IrtAB imports and reduces mycobactins, but contradictorily it assumes an ABC exporter fold (Arnold, Weber, Gonda et al., Nature, 2020). In this new study, we used extensive cryo-EM analysis and DEER measurements to explore the conformational landscape of IrtAB and determine the molecular mechanism of mycobactin translocation along the inner membrane. Most interestingly, we found that instead of a three-state, ABC exporter-like system, IrtAB alternates between 2 states: an inward-facing and outward-occluded conformations, and does not adopt a fully outward-facing state.
In one of our cryo-EM structures, we were able to capture mycobactin translocation along the lipid bilayer, using membrane mimicking nanodiscs. Here, mycobactin is wedged into a unique lateral crevice, that is only formed in the outward-occluded state. Mutational analyses along this unique crevice and in vivo uptake assays show that structurally disrupting this crevice abolishes mycobactin transport. The combined structural and functional characterization of the crevice suggests that mycobactins are likely imported in a “credit-card swipe” manner: the lipid tail remains in the membrane while the iron-chelating head group slides along the crevice.
Notably, we identified a zinc-binding site along this crevice, facing the cytosol. This zinc binding was proven to be essential for transport function. Interestingly, while ATP hydrolysis still occurs when this site is disrupted, iron transport does not - highlighting zinc’s critical role in coupling energy use to substrate import. This finding is particularly relevant considering that during infection, M. tuberculosis encounters highly variable Zn²⁺/Fe³⁺ ratios. In macrophage vacuoles, where mycobacteria may reside, metal ion concentrations can shift rapidly, where IrtAB may play a role in adapting faster to these changing conditions than transcriptional level mechanisms.
Our structural and mechanistic insights lay the groundwork and provide useful tools for future structure-based drug development targeting IrtAB or the broader mycobactin-mediated iron acquisition system.
Dr Galo A. Goig et al.
Transmission as a Key Driver of Resistance to the New Tuberculosis Drugs
 Multidrug-resistant tuberculosis (MDR-TB) is a major public health concern. Recently, the World Health Organization endorsed new regimens based on novel and repurposed drugs (i.e bedaquiline, pretomanid, and linezolid with or without moxifloxacin [BPaL/M]). These new regimens have proved highly effective against MDR-TB, bringing new hope in the fight against the disease. However, despite the recent introduction of these regimens, emergence of resistance to these new therapeutic agents has already been reported.
Multidrug-resistant tuberculosis (MDR-TB) is a major public health concern. Recently, the World Health Organization endorsed new regimens based on novel and repurposed drugs (i.e bedaquiline, pretomanid, and linezolid with or without moxifloxacin [BPaL/M]). These new regimens have proved highly effective against MDR-TB, bringing new hope in the fight against the disease. However, despite the recent introduction of these regimens, emergence of resistance to these new therapeutic agents has already been reported.
In our study published in The New England Journal of Medicine, we aimed to better understand the global emergence of M. tuberculosis strains resistant to these new antibiotics, and particularly how patient-to-patient transmission contributes to the spread of these highly drug-resistant strains.
First, we analyzed the genomes of a 13-year, nationwide collection of nearly 7,000 M. tuberculosis isolates from Georgia, a country with a high incidence of MDR-TB. We identified 60 highly drug-resistant strains that carried mutations known to confer resistance to first-line drugs (i.e rifampicin and isoniazid) along with fluoroquinolones and at least one of the BPaL compounds. By comparing the genomes of the bacteria and analyzing the specific drug resistance-conferring mutations, we identified transmission chains in which these highly drug-resistant strains were likely transmitted from patient-to-patient. We estimated that 28% of all highly drug-resistant cases in Georgia were involved in transmission chains.
Next, we expanded our analysis to over 81,000 M. tuberculosis genomes from global sources, identifying 454 additional highly drug-resistant strains across 26 countries. Our genomic analysis revealed that 28% of these strains, from 10 countries spanning the Americas, Europe, Africa and Asia, were involved in transmission chains. Additionally, we identified 9 M. tuberculosis isolates with mutations conferring resistance to all BPaL/M compounds, therefore potentially classifying as totally drug-resistant.
Our results show that despite the recent introduction of BPaL/M, resistance to these new drugs is already emerging and spreading worldwide. These findings call for urgent improvements in diagnostic capacity, infection control, and surveillance, without which the longevity of BPaL/M is at risk.
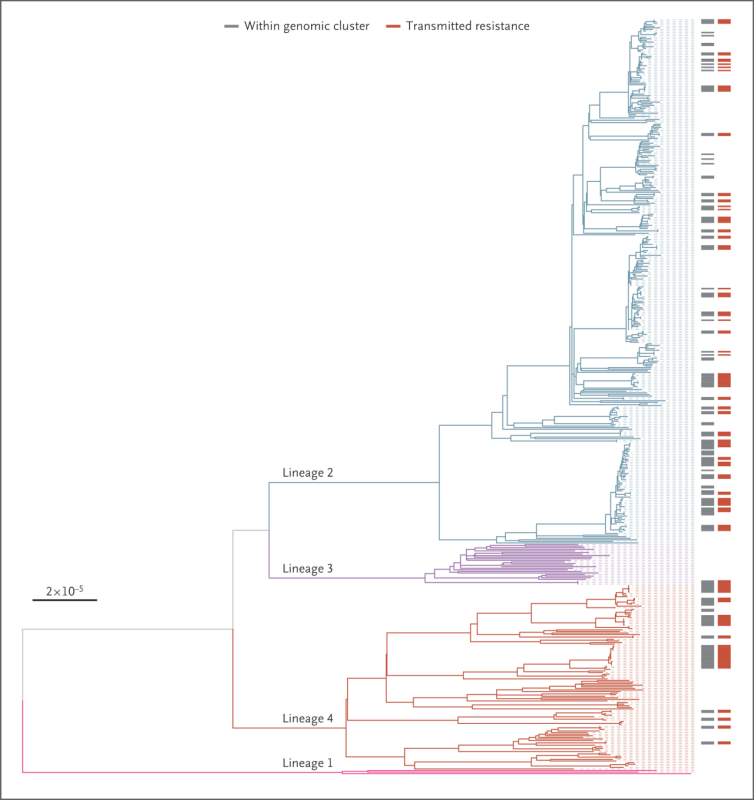
Figure legend:
Maximum-Likelihood Phylogeny of the Identified Highly Drug-Resistant Mycobacterium tuberculosis Strains.
Branches are colored according to the M. tuberculosis complex lineage: pink for lineage 1, blue for lineage 2, purple for lineage 3, and red for lineage 4. Discrete heat maps next to the phylogeny indicate whether the strains were within a genomic cluster (gray) and whether they shared the same drug resistance–conferring mutations, in which case transmitted resistance was inferred (red). The scale bar indicates the number of substitutions per site per genome.
